Cells, the basic units of biological structure and function, vary broadly in type and state. In most biological systems, our knowledge of cellular diversity is incomplete, as for complex tissues like the nervous system (brain cells).The characterization of single-cell identity and function, as the understanding of the functional capacities and responses of each cell type, will accelerate biological discoveries. It could be for cancer, for tumors, for almost anything that is likely to have diversity in the population of cells. However, today’s techniques do not present an easy enough way to analyze a large amount of individual cells simultaneously.
In this way, fast, scalable approaches are needed to characterize complex tissues with many cell types and states.
PRINCIPLE & BENEFITS OF DROP-SEQ
Drop-Seq is a strategy based on the use of microfluidics for quickly profiling thousands of individual cells simultaneously by encapsulating them in tiny droplets for parallel analysis.
These nanoliter-scale aqueous compartments have been used for many applications in microfluidic devices: nanoparticles fabrication, emulsions and foams, drug delivery, but also as tiny reaction chambers for PCR and reverse transcription [3]. Drop-Seq uses droplets to compartmentalize cells into nanoliter-sized reaction chambers for analysis of their mRNA transcripts while remembering transcripts’ cell of origin, using a molecular barcoding strategy. With this technique, one single scientist can make 10 000 single cell libraries, every day, in parallel inexpensive and easy experiments. This method will thus allow the creation of molecular atlases of gene expression for known cell classes and novel candidate cell subtypes.
Drop-seq takes profit of the advantages of microfluidics being:
high throughput results in a short period of time
minimize consumption of expensive samples
DROP-SEQ CONSISTS OF THE FOLLOWING STEPS
Prepare a single-cell suspension from a tissue
Prepare the barcoded primers (either at the surface of the microparticles, either inside)
Use a microfluidic device to co-encapsulate each cell individually with a distinctly barcoded microparticle in a tiny droplet
Once isolated in droplets, lyse the cells, which release their mRNAs, which then hybridize to the primers
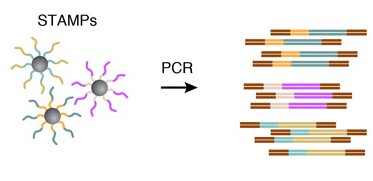
Break the droplets and generate STAMPs (single-cell transcriptomes attached to microparticles)
Amplify the STAMPs
Sequencing and analysis: use of the STAMP barcodes to infer each transcript’s cell of origin
2 types of beads can be used for Drop-Seq:
“Simple” microparticles
Hydrogel microparticles
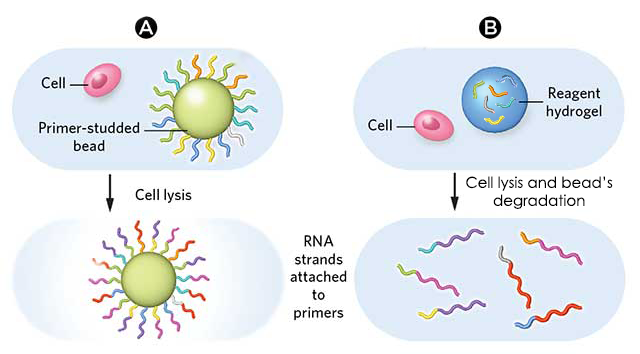
This work focuses on the use of “simple” microparticles and addresses briefly hydrogel microparticles, for which the principle stays the same.DROP-SEQ USING SIMPLE MICROPARTICLES
1. PREPARE A SINGLE-CELL SUSPENSION FROM A COMPLEX TISSUE
A complex tissue is dissociated into individual cells.
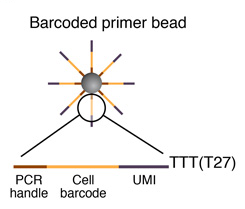
2. PRIMER SYNTHESIS
Sequence of primers on the microparticle
Each microparticle contains more than 108 individual primers that share the same “PCR handle” and “cell barcode”, but have different unique molecular identifiers (UMIs). PCR handle is in fact a constant sequence identical on all primers and beads, which allows PCR amplification after STAMP formation. Cell barcode, only identical across all the primers of a same microparticle but different from cell barcodes on other beads, allows to recover the cells’ origin. UMIs, different on each primer, allow mRNA transcripts to be digitally counted and to identify PCR duplicates. Finally, a 30-bp oligodT sequence is present at the end of all primer sequences for capturing mRNAs and priming reverse transcription.
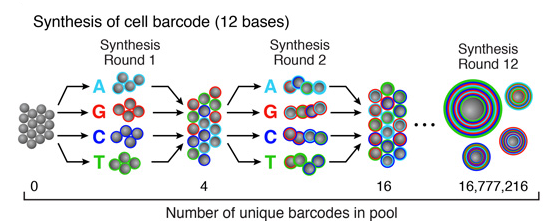
Split-and-pool synthesis of the cell barcode
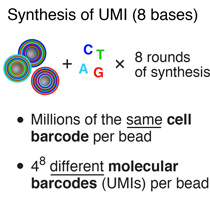
To generate the cell barcode, the pool of microparticles is repeatedly split into four equally sized oligonucleotide synthesis reactions, to which one of the four DNA bases is added. The microparticles are then pooled together after each cycle, and undergo a total of 12 split-pool cycles. The result is a pool of microparticles, each possessing one of 412 (16,777,216) possible sequences of DNA bases.Synthesis of a unique molecular identifier (UMI)
The synthesis of UMIs takes place after the completion of the “split-and-pool” synthesis cycles. All microparticles are subjected together to eight rounds of degenerate synthesis with all four DNA bases available during each cycle, such that each individual primer receives one of 48 (65,536) possible sequences (UMIs).
3. MICROFLUIDIC DEVICE
Once the single-cell suspension and the microparticles ready, individual cells are encapsulated in droplets together with microparticles, using a custom-designed microfluidic device.
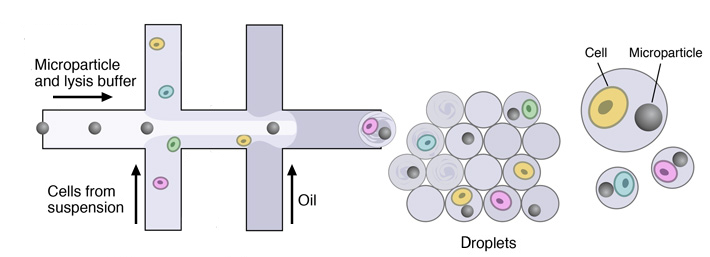
This device joins two aqueous flows before their compartmentalization into discrete droplets. Laminar flow prevents mixing of the two aqueous inputs prior to droplet formation. One flow contains cells, and the other flow contains barcoded primer beads suspended in a lysis buffer.
MICROFLUIDIC SETUP
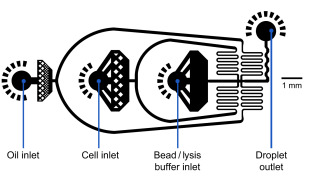
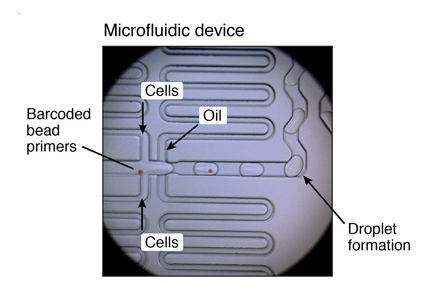
Flow rates
Oil: 15,000 µl/hr = 250 µl/min
Cells: 4,000 µl/hr ~ 66,7 µl/min
Beads: 4,000 µl/hr ~ 66,7 µl/minExample of a microfluidic device for Drop-Seq
List of components
An inverted microscope
Pressure controller + 3 Flow sensor/ Three syringe pumps
3 falcon tubes/ 3 mL syringes
Magnetic mixing system
Microfluidic tubing for the fluidic connection of the setup elements
Microfluidic fittings & connectors
PDMS co-flow microfluidic droplet generation device
100 micron cell strainers for beads
40 micron cell strainers for cells
Counting chamber
Set-up Diagram
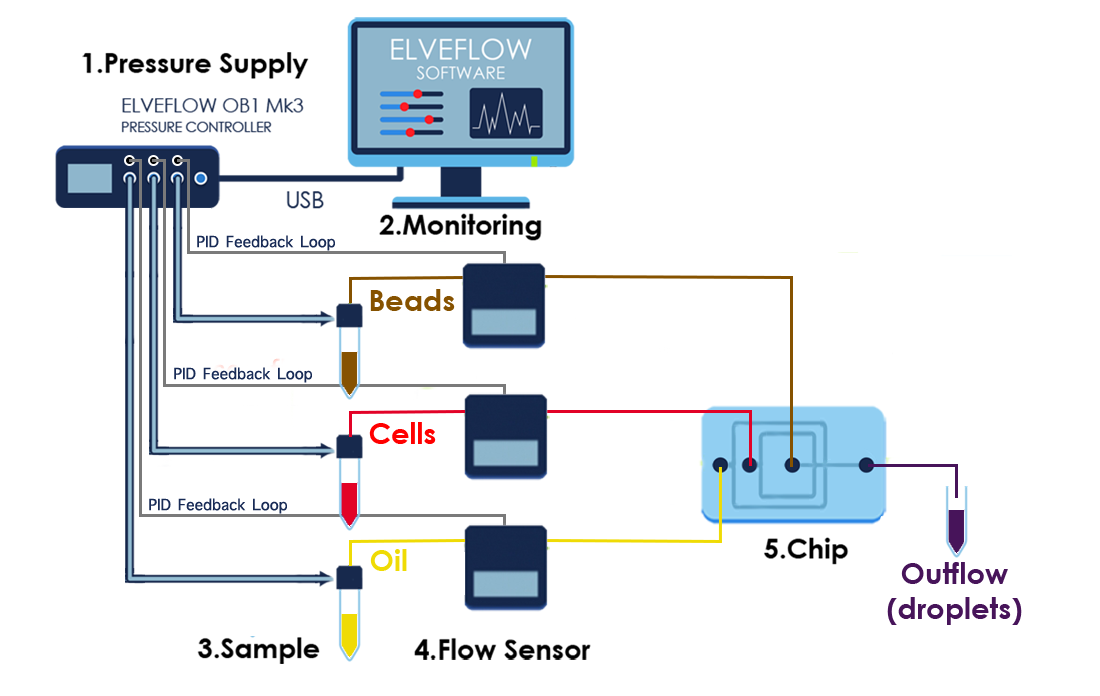
4. CELL LYSIS AND RNA HYBRIDIZATION
Immediately following droplet formation, each cell is lysed within the droplet and its mRNAs are released. Then they hybridize to the primers on its companion microparticle surface.
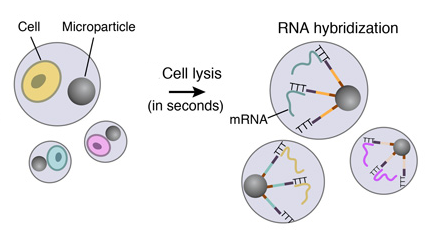
5. STAMPS GENERATION
To efficiently generate thousands of STAMPs at once, the droplets are broken by adding a reagent to destabilize the oil-water interface, and the microparticles are collected and washed. The mRNAs are then reverse-transcribed into cDNAs together in one reaction, forming a set of beads called “single-cell transcriptomes attached to microparticles” (STAMPs).
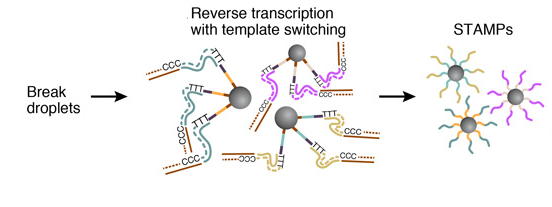
6. AMPLIFICATION OF STAMPS
The barcoded STAMPs can then be amplified in pools by PCR reaction for high-throughput mRNA-sequencing, to analyze any desired number of individual cells.
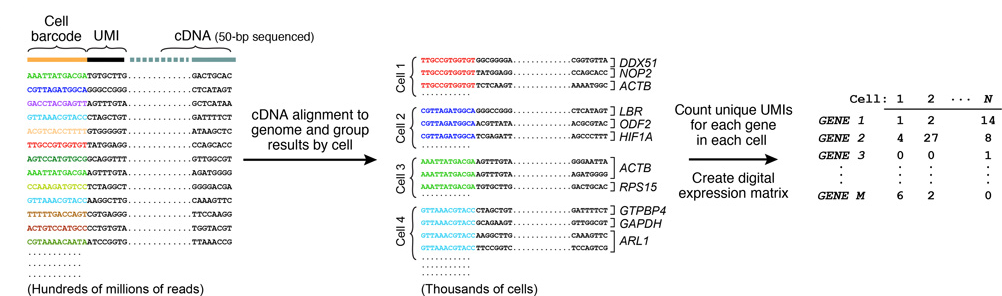
7. SEQUENCING AND ANALYSIS
Resulting molecules are sequenced from each end using high-capacity parallel sequencing. The reads are first aligned to a reference genome to identify the gene-of-origin of the cDNA. Next, reads are organized by their cell barcodes, and the number of mRNA transcripts of each gene ascertained in each cell is digitally counted. This is where UMIs come into play, avoiding double-counting sequence reads from the same mRNA transcript. A matrix of digital gene-expression measurements (one measurement per gene per cell) can then be established for further analysis.
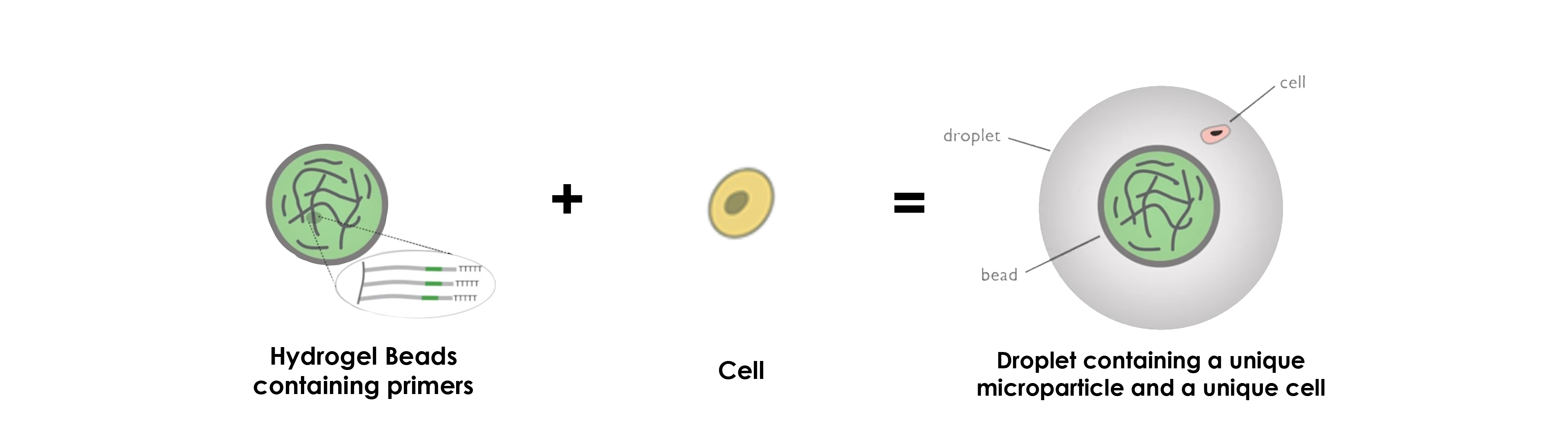
DROP-SEQ USING HYDROGEL MICROPARTICLES
Concerning the use of hydrogel beads, the operating principle stays more or less the same, namely that the greatest difference concerns the primers, which are inside the microparticles instead of at their surface.
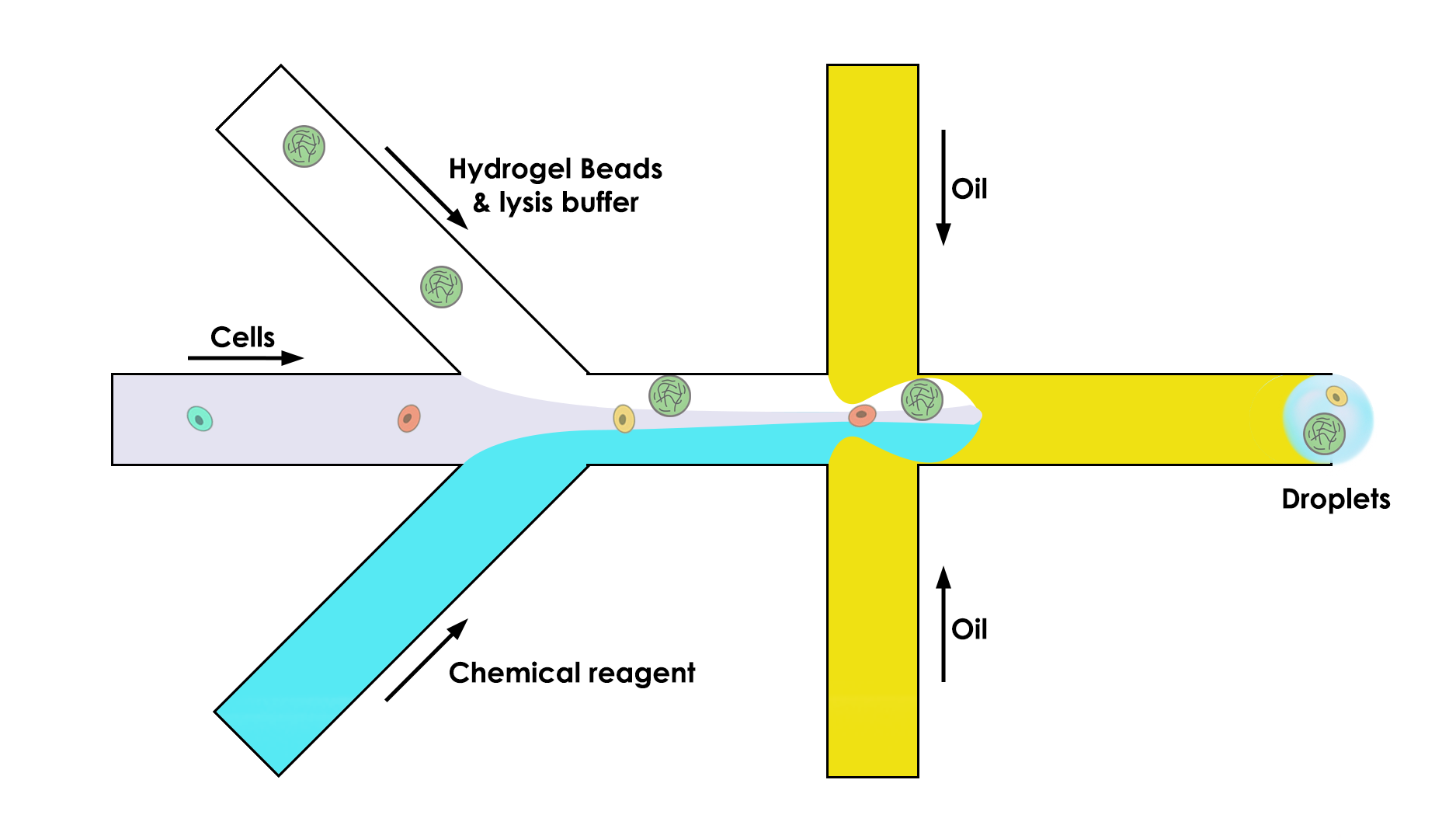
To do so, the microfluidic device is composed of three channels instead of two:
One that brings the bead (the key of this whole process)
One that brings the cell in the droplet
One that brings the chemical reagent that we need to do the analysis
As for the use of “simple microparticles”, hydrogel beads contain primers that can be used for a reverse transcription reaction, which will then subsequently barcode the contents of the cells inside the droplet. A collection of DNA sequences that are copies of the RNA sequences are thus obtained, and these sequences are now sorted by what drop they came from.
Then, it is possible to break the droplets and treat the entire cell population as a bulk sample, knowing that each of the cells has been individually barcoded.